Welcome
SMARTEn (System for Mobile Analysis in Real-Time of ENvironment)
SMARTEn aims to provide new ways to analyze metagenomic DNA using fully mobile setups. The project is run by our interdisciplinary team of experts in computer science and environmental engineering.
Our codebase is open source and free and available from our official GitLab repository. Here you can find out more about software tools we are developing.
SKiM
SKiM is a memory-efficient tool for classifying ONT reads in large metagenomic datasets. By using syncmers alongside advanced data compression and statistical correction techniques, SKiM enables fast, accurate classification across large-scale DNA reference databases while drastically reducing memory consumption.
To download SKiM visit the official SKiM GitLab repository.
Coriolis
Coriolis is a fully functional metagenomic classifier designed for lightweight mobile devices, such as ONT’s MinIT or the latest MinION sequencers. Similar to Centrifuge, Coriolis’s classification algorithm is based on exact-match queries of arbitrary lengths. Through the use of optimized external memory data structures, Coriolis performs optimally even in extremely low-memory environments. Coriolis is designed to work with mobile devices, and supports both Intel and ARM processors. It generates outputs that are 100% compatible with Centrifuge, and hence can be used as a drop-in replacement in the existing workflows.
To download Coriolis visit the official Coriolis GitLab repository.
Checkout out this demo of coriolis 0.1.2 running in real-time alongside dorado 0.6.2 basecaller. The experiment is running on an NVIDIA Jetson Orin Nano with 8GB of RAM and 4TB of SSD (WD_BLACK 4TB SN750 NVMe). Coriolis uses 1GB of RAM to cache the reference database of NCBI RefSeq complete genomes of all archaea, bacteria and viruses (over 150GB of raw DNA sequences). The performance is achieved by using a combination of the SMARTEn runtime and our Compacted String B-Tree data structure. In this experiment, Coriolis is able to run at speeds close to 50 reads/s. For smaller reference databases (yet still exceeding all available RAM), we were able to achieve performance of up to 128 reads/s.
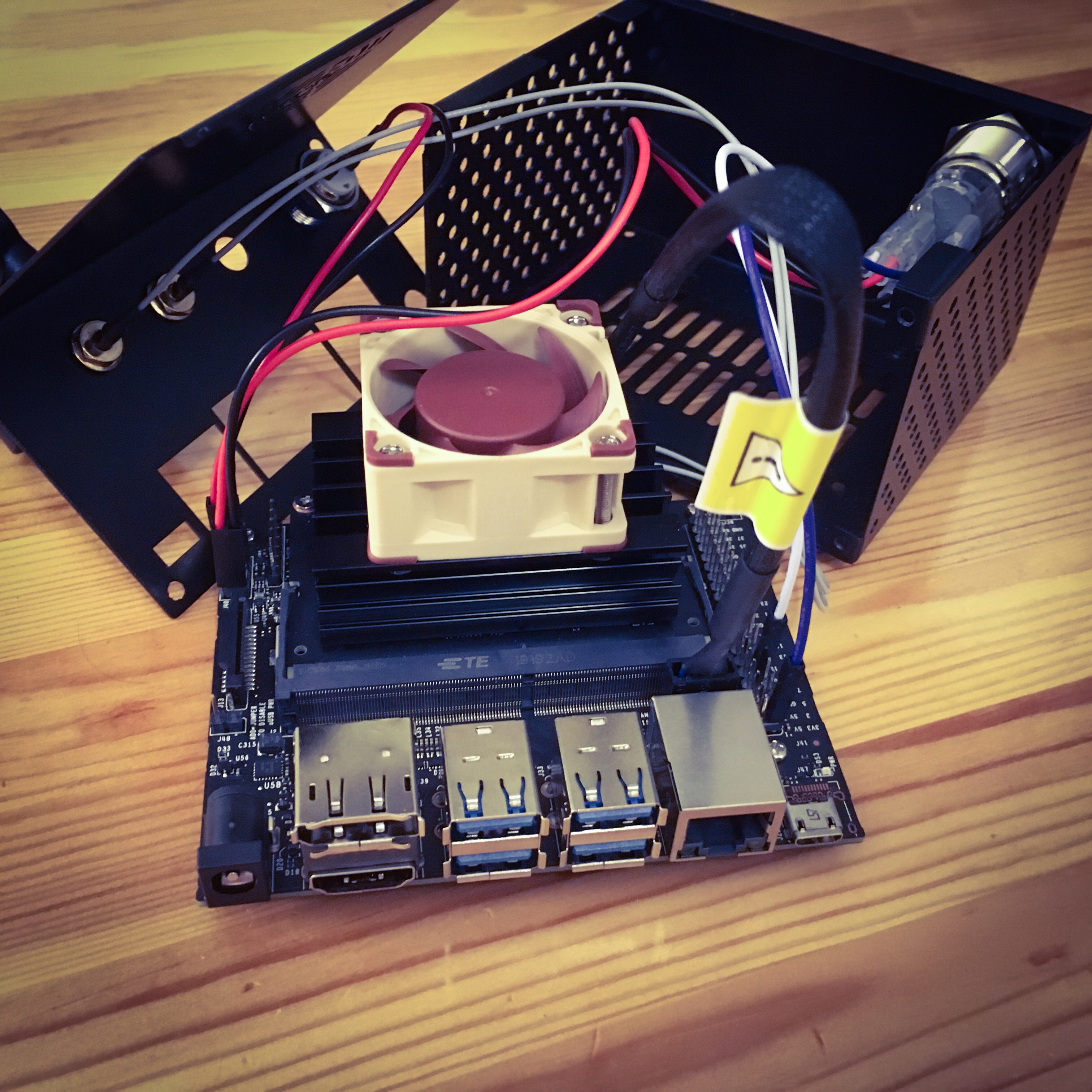
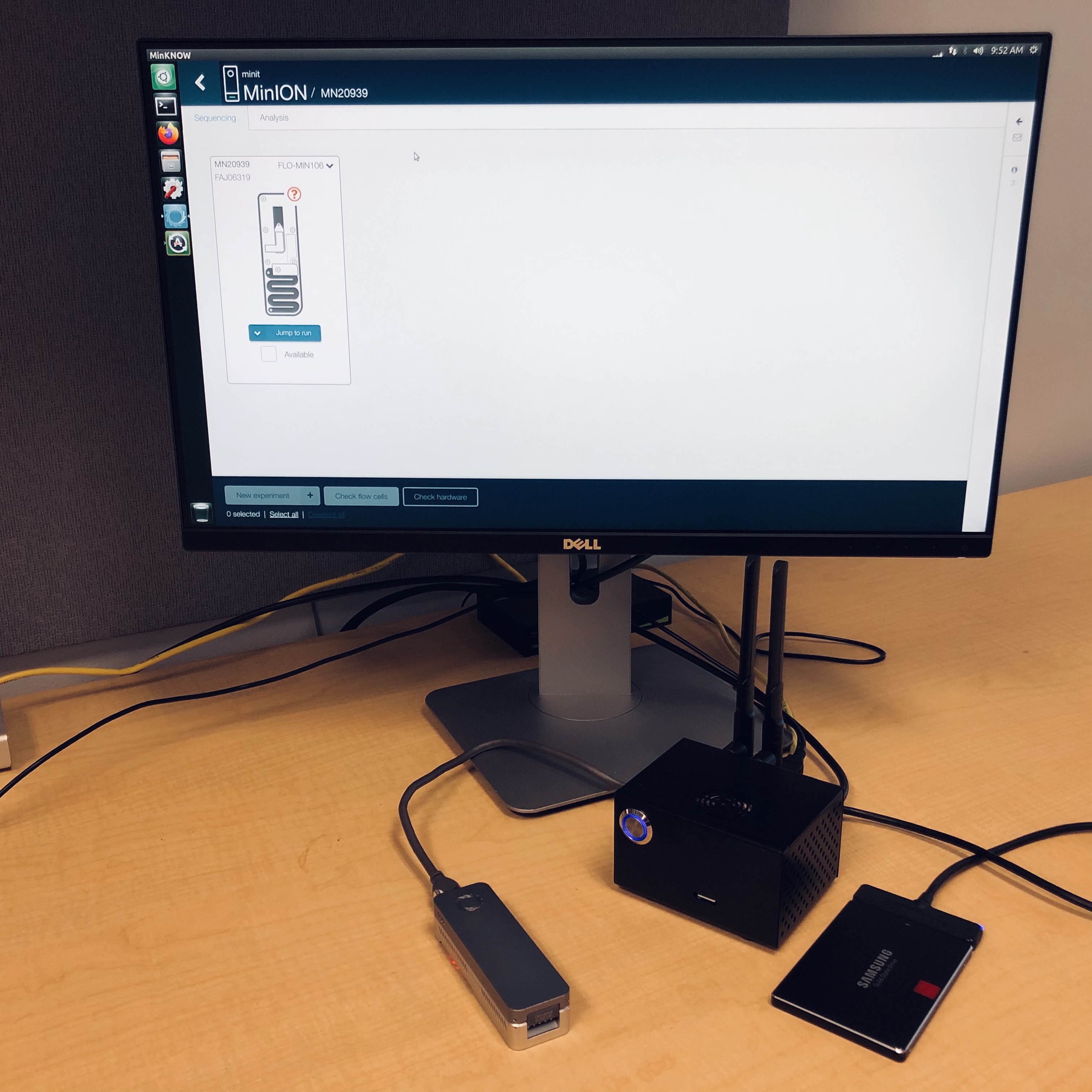
Our Hardware
To run the project we are using two key pieces of technology. The first one is MinION sequencer from Oxford Nanopore Technology. The MinIONs are the enabling technology behind portable in the field metagenomics. The pictures below show one of the three devices we are using in our labs.


The second component are various models of a Supercomputer On a Chip (SCoC). The specific platforms we are using are based on NVIDIA’s Jetson modules. We have been using NVIDIA Jetson TX1, NVIDIA Jetson Nano and more recently NVIDIA Jetson Orin Nano. While these boards come with impressive compute power (delivered by the GPGPU cores) they have limited main memory. This prompted us to develop Coriolis.
Below we provide example pictures of our setup.


Emulating MinION
The mGRUE team of undergraduate students under the supervision of Dr. Kris Schindler built a hardware/software combo that allows us to emulate MinION sequencer. You can learn more about the project from their repository. This project was done as part of the fantastic CSE 453 “Hardware/Software Integrated Systems Design 2” course at UB.
The mGRUE team are:
- Jonathan Baffo
- Warren Green
- Christian Palladino
- Brian Scorcia
- Aaron Siegel
- Lucas Simpson
References
- T. Schneggenburger, J. Zola SKiM: Accurately Classifying Metagenomic ONT Reads in Limited Memory, Bioinformatics 2025.
- A.J. Mikalsen, J. Zola, Coriolis: Enabling Metagenomic Classification on Lightweight Mobile Devices, ISMB 2023.
- V. Zheng, A.E. Sariyuce, J. Zola, Identifying Taxonomic Units in Metagenomic DNA Streams on Mobile Devices, TCBB 2022.
- V. Zheng, A.E. Sariyuce, J. Zola, Identifying Taxonomic Units in Metagenomic DNA Streams, BioKDD 2020.
- S. Ko, L. Sassoubre, J. Zola, Applications and Challenges of Real-time Mobile DNA Analysis, HotMobile 2018.