Computer
Assisted Surgery
CADI has developed an image guided
neurosurgery toolkit to produce optimum plans resulting in
minimally invasive surgeries. The Computer Assisted Surgery
(CAS) engine covers several research and engineering
solutions.
Finite Element Modeling (FEM) to predict
brain shift:
FEM is used to predict intraoperative brain shift during
neurosurgery; the system uses a three-dimensional (3D)
patient-specific finite element (FE) brain model with
detailed anatomical structures using quadrilateral and
hexahedral elements.
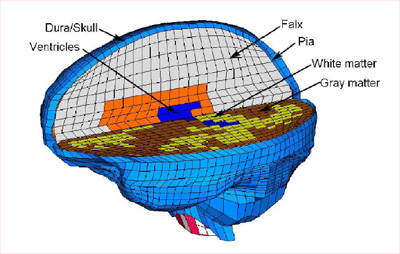
Methods: A template-based algorithm was developed to build a
3D patient-specific FE brain model. The template model is a
50th percentile male FE brain model with gray and white
matter, ventricles, pia mater, dura mater, falx cerebri,
tentorium cerebelli, brainstem and cerebellum. Two patient
specific models were constructed to demonstrate the
robustness of this method. Gravity-induced brain shift after
dura opening was simulated based on one clinical case of
computer assisted neurosurgery for model validation. The
pre-operative MR images were updated by the FE results, and
displayed as intraoperative MR images easily recognizable by
surgeons.
A set of algorithms for developing a 3D patient-specific FE
brain model have been developed. Gravity-induced brain shift
can be predicted by this model and displayed as high
resolution MR images. Such strategy can be used for not only
intraoperative MRI updating, but also pre-surgical planning.
Wireless
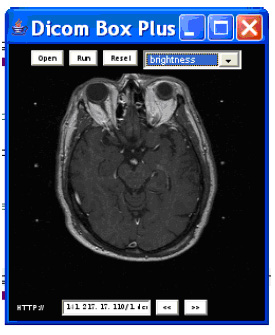
We developed DICOMBox tool based on the DICOM processing
algorithm in Eview project, which can view and edit the
Dicom images on hand held devices. This work shows the
promising future to move computing non-intensive
functionalities of the CAS Engine to hand held platform. In
terms of the secure access for the CAS Engine, a location
based access control model is proposed as a comprehensive
solution for CAS Engine to meet the HIPAA standard.
Database
The CAS Database set up in a secure Client/Server
architecture allows users to upload case information, image
data, planning and annotation information. The system
supports several types of navigational queries that assist a
surgeon in decision making.
Identify/design and develop advanced (3D)
interfaces for navigational queries
The surgical interface will also allow users to navigate
possible surgical trajectory even before entering the OR.
This is accomplished using a new indexing structure
developed by over the course of the CAS program. Called the
target tree, this index is a variable height tree that
recursively decomposes the search space around a single
target point. The index allows for insertion and deletion
operations to be intermixed with searches. The target point
of the index is the end goal of a surgical procedure,
usually a tumor that must be removed.
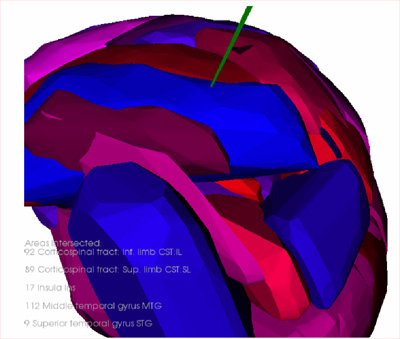
Augmented Reality
We have successfully developed and implemented a prototype
for Augmented Reality (AR) system to visualize invisible
critical structures of brain in the real view of patient
phantom.
Landmark-based Patient & Atlas
Co-Registration
The transfer of anatomical knowledge from 3D atlases to
patient images via image-atlas co-registration is a very
helpful tool in applications such as diagnosis, therapy
planning, and simulation. However, there are anatomical
differences among individual patients that make registration
difficult; accurate voxel-wise fusion of different
individuals is an open problem. For planning and simulation
applications accuracy is essential, because any geometrical
deviation may be harmful to a patient.
Landmarks-based registration is one of the most popular
algorithms in atlas-based application. We have implemented
landmarks based registration as our first atlas registration
algorithm. Here, AC, PC, L, and R were chosen as our control
points.
CADI group has worked on mainly five rigid
registration algorithms and a deformable registration
technique. Following are the registration techniques:
- Multi-Resolution Mutual Information
- Mutual Information
- Landmark based rigid registration
- Landmark with Mutual Information.
The concentration has been to achieve best
results with minimal time take for registration or fusion of
mutli-modality data.
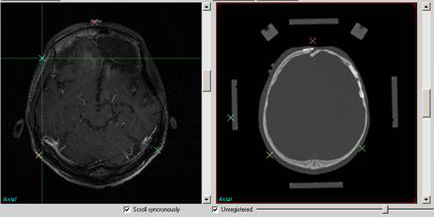
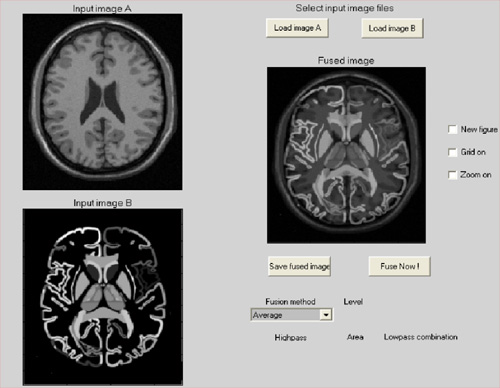
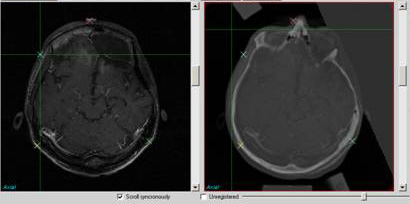
Figure shows the registration result using
algorithm “Landmark + Mutual information” and a simple image
fusion.
(top) |



