Research
Funding:
-
 DOE IAV
DOE IAV
-
 NSF SaTC
NSF SaTC
-
 NSF DIBBS
NSF DIBBS
-
 NSF SATC
NSF SATC
-
 AMAZON
AMAZON
-
 MSFT
MSFT
Insider Threat Detection - Exploiting Data Relationships to Detect Insider Attacks
Insider attacks present an extremely serious, pervasive and costly security problem under critical domains such as national defense and financial and banking sector. Accurate insider threat detection has proved to be a very challenging problem. This project explores detecting insider threats in a banking environment by analyzing database searches. This research addresses the challenge by formulating and devising machine learning-based solutions to the insider attack problem on relational database management systems (RDBMS), which are ubiquitous and are highly susceptible to insider attacks. In particular, the research uses a new general model for database provenance, which captures both the data values accessed or modified by a user's activity and summarizes the computational path and the underlying relationship between those data values. The provenance model leads naturally to a way to model user activities by labeled hypergraph distributions and by a Markov network whose factors represent the data relationships. The key tradeoff being studied theoretically is between the expressivity and the complexity of the provenance model. The research results are validated and evaluated by intimately collaborating with a large financial institution to build a prototype insider threat detection engine operating on its existing operational RDBMS. In particular, with the help of the security team from the financial institution, the research team addresses database performance, learning scalability, and software tool development issues arising during the evaluation and deployment of the system.
This research addresses the challenge by formulating and devising machine learning-based solutions to the insider attack problem on relational database management systems (RDBMS), which are ubiquitous and are highly susceptible to insider attacks. In particular, the research uses a new general model for database provenance, which captures both the data values accessed or modified by a user's activity and summarizes the computational path and the underlying relationship between those data values. The provenance model leads naturally to a way to model user activities by labeled hypergraph distributions and by a Markov network whose factors represent the data relationships. The key tradeoff being studied theoretically is between the expressivity and the complexity of the provenance model. The research results are validated and evaluated by intimately collaborating with a large financial institution to build a prototype insider threat detection engine operating on its existing operational RDBMS. In particular, with the help of the security team from the financial institution, the research team addresses database performance, learning scalability, and software tool development issues arising during the evaluation and deployment of the system.
Understanding Rumor Propagation in Social Networks
One prominent threat action (attack vector) in social cyber-attacks is the use of rumors. We are developing methods that integrate social science and computer science methods and new analysis techniques to process large-scale tweet data for detection and alerting of rumors. One of the key component of such research is the availability of a testbed for validating methods. In the context of social media, this requires a realistic network structure and models for simulating the spread of information, including rumors. We have developed statistical generative models for graphs that are able to capture the properties of graphs/networks that occur in real world [IEEE BigData, 2015]. While in the past, focus was on generating graphs which follow general laws, such as the power law for degree distribution, current models have the ability to learn from observed graphs and generate synthetic approximations. The primary emphasis of existing models has been to closely match different properties of a single observed graph. Such models, though stochastic, tend to generate samples which do not have significant variance in terms of the various graph properties. We argue that in many cases real graphs are part of a population (e.g., networks sampled at various time points, social networks for individual schools, healthcare networks for different geographic regions, etc.). Graphs in a population exhibit variance. However, existing models are not designed to model this variance, which could lead to issues such as overfitting. We propose a graph generative model that focuses on matching the properties of real graphs and the natural variance expected for the corresponding population. The proposed model adopts a mixture-model strategy to expand the expressiveness of Kronecker product based graph models (KPGM), while building upon the two strengths of KPGM, viz., ability to model several key properties of graphs and to scale to massive graph sizes using its elegant fractal growth based formulation. The proposed model, called x-Kronecker Product Graph Model, or xKPGM, allows scalable learning from observed graphs and generates samples that match the mean and variance of several salient graph properties.
While in the past, focus was on generating graphs which follow general laws, such as the power law for degree distribution, current models have the ability to learn from observed graphs and generate synthetic approximations. The primary emphasis of existing models has been to closely match different properties of a single observed graph. Such models, though stochastic, tend to generate samples which do not have significant variance in terms of the various graph properties. We argue that in many cases real graphs are part of a population (e.g., networks sampled at various time points, social networks for individual schools, healthcare networks for different geographic regions, etc.). Graphs in a population exhibit variance. However, existing models are not designed to model this variance, which could lead to issues such as overfitting. We propose a graph generative model that focuses on matching the properties of real graphs and the natural variance expected for the corresponding population. The proposed model adopts a mixture-model strategy to expand the expressiveness of Kronecker product based graph models (KPGM), while building upon the two strengths of KPGM, viz., ability to model several key properties of graphs and to scale to massive graph sizes using its elegant fractal growth based formulation. The proposed model, called x-Kronecker Product Graph Model, or xKPGM, allows scalable learning from observed graphs and generates samples that match the mean and variance of several salient graph properties.
Identifying Fraud, Waste, and Abuse from Massive Healthcare Claims Data
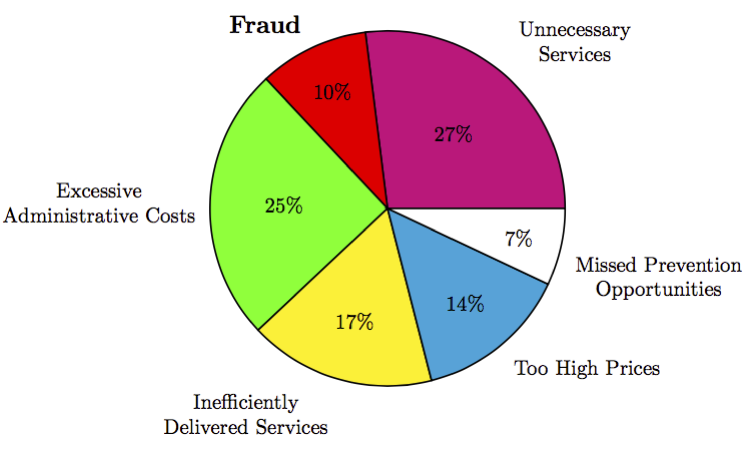 Healthcare spending in United States is one of the key issues targeted by policy makers, owing to the fact that healthcare spending in US is a major contributor to the high national debt levels that are projected for next two decades. In 2008, the total healthcare spending in US was 15.2% of its GDP (highest in the world) and is expected to reach as much as 19.5% by 2017. But while the healthcare costs have risen (by as much as 131% in the past decade), the quality of healthcare in the US has not seen comparable improvements. We explore the promise of big data analytics to leverage the massive healthcare claims data for better models for preventing healthcare fraud, waste and abuse. We translate the problem of analyzing healthcare data into some of the most well known analysis problems in the data mining community, social network analysis, text mining, and temporal analysis and higher order feature construction, and describe how advances within each of these areas can be leveraged to understand the domain of healthcare.
Healthcare spending in United States is one of the key issues targeted by policy makers, owing to the fact that healthcare spending in US is a major contributor to the high national debt levels that are projected for next two decades. In 2008, the total healthcare spending in US was 15.2% of its GDP (highest in the world) and is expected to reach as much as 19.5% by 2017. But while the healthcare costs have risen (by as much as 131% in the past decade), the quality of healthcare in the US has not seen comparable improvements. We explore the promise of big data analytics to leverage the massive healthcare claims data for better models for preventing healthcare fraud, waste and abuse. We translate the problem of analyzing healthcare data into some of the most well known analysis problems in the data mining community, social network analysis, text mining, and temporal analysis and higher order feature construction, and describe how advances within each of these areas can be leveraged to understand the domain of healthcare.
Probabilistic Models for Precision Medicine (Deep Phenotyping)
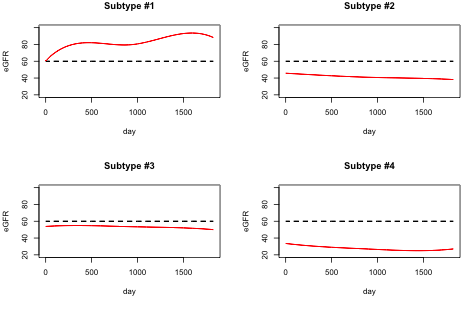 Precision medicine is an emerging medicine paradigm that takes into account individual variability in genes, environment, and lifestyle to come up with more effective treatment and prevention strategies. The recently launched Precision Medicine Initiative underscores the importance of this area. One of the key scientific driver in the context of precision medicine is the ability to identify stratified subgroups of individuals who exhibit similar disease related behavior. In medical terms, this process is known as phenotyping, where the phenotypes are the observable traits exhibited by the subpopulation. Precision medicine demands identification of precise phenotypes, also referred to as deep phenotyping. In this research we are focusing on extracting deep phenotypes from clinical data for patients suffering from Chronic Kidney Disease (CKD).
Precision medicine is an emerging medicine paradigm that takes into account individual variability in genes, environment, and lifestyle to come up with more effective treatment and prevention strategies. The recently launched Precision Medicine Initiative underscores the importance of this area. One of the key scientific driver in the context of precision medicine is the ability to identify stratified subgroups of individuals who exhibit similar disease related behavior. In medical terms, this process is known as phenotyping, where the phenotypes are the observable traits exhibited by the subpopulation. Precision medicine demands identification of precise phenotypes, also referred to as deep phenotyping. In this research we are focusing on extracting deep phenotypes from clinical data for patients suffering from Chronic Kidney Disease (CKD).
Improving Quality of Health Care using Tree-structured Sparsity Norms
 Hospital readmissions have become one of the key measures of healthcare quality. Preventable readmissions have been identified as one of the primary targets for reducing costs and improving healthcare delivery. However, most data driven studies for understanding readmissions have produced black box classification and predictive models with moderate performance, which precludes them from being used effectively within the decision support systems in the hospitals. In this research we exploit the hierarchical relationships between diseases via structured sparsity-inducing norms for predicting readmission risk for patients based on their disease history and demographics.
Hospital readmissions have become one of the key measures of healthcare quality. Preventable readmissions have been identified as one of the primary targets for reducing costs and improving healthcare delivery. However, most data driven studies for understanding readmissions have produced black box classification and predictive models with moderate performance, which precludes them from being used effectively within the decision support systems in the hospitals. In this research we exploit the hierarchical relationships between diseases via structured sparsity-inducing norms for predicting readmission risk for patients based on their disease history and demographics.
Scaling Gaussian Process Models for Big Scientific Data
 Gaussian process regression (GPR) based models, also known as Kriging in geostatistics, have been widely used in the area of geostatistics. Lately, such methods have found renewed interest in areas such as Uncertainty Quantification for large scale scientific simulations. While Gaussian process formulation has several attractive properties including ability to simultaneously model spatial, temporal, and functional dependencies, the inherent computational complexity associated with the analysis often prevents their application to large scale studies. Our research focuses on scaling GPR models for applications such as biomass monitoring and uncertainty quantification in large scale scientific simulation.
We have developed a suite of scalable algorithms that can be deployed on a range of High Performance Computing (HPC) infrastructures such as traditional clusters (using both MPI and Map-reduce based implementations) and GPU accelerated nodes.
Gaussian process regression (GPR) based models, also known as Kriging in geostatistics, have been widely used in the area of geostatistics. Lately, such methods have found renewed interest in areas such as Uncertainty Quantification for large scale scientific simulations. While Gaussian process formulation has several attractive properties including ability to simultaneously model spatial, temporal, and functional dependencies, the inherent computational complexity associated with the analysis often prevents their application to large scale studies. Our research focuses on scaling GPR models for applications such as biomass monitoring and uncertainty quantification in large scale scientific simulation.
We have developed a suite of scalable algorithms that can be deployed on a range of High Performance Computing (HPC) infrastructures such as traditional clusters (using both MPI and Map-reduce based implementations) and GPU accelerated nodes.
iGlobe/webGlobe: Interactive Visualization and Analysis of Geospatial Data Sets on a 3D Globe

iGlobe is a unified framework that allows integration of disparate geospatial data sources that include remotely sensed observation data, climate model outputs, population and critical infrastructure data, and provide an interface for both client visual analytics and the sever-side data mining capabilities. We see several benefits of such unified framework for both scientists and policy makers. First, it hides complex transformations that are required to overlay climate variables on geospatial data layers like land use and land cover. Second, it allows visual analytics capabilities where users can directly observe and correlate disparate events, for example, proximity of temperature extremes to the population and precipitation extremes over croplands. Thus the tool is invaluable to scientists, policy makers and general public alike.
The webGlobe project combines the rich capabilities of Web World Wind with the NetCDF data access capabilities of iGlobe. Just like Web World Wind, webGlobe runs on all major operating systems, desktop and mobile devices, and web browsers. The video below demonstrates how webGlobe allows access and visualization of remotely available NetCDF data. However, the key capability of webGlobe is under the hood. To allow visualization and analysis of a wide variety of geospatial data sets, webGlobe has a decoupled client-server architecture.