This is an old revision of the document!
SMARTEn
System for Mobile Analysis in Real-Time of ENvironment
This project is supported by the National Science Foundation under the award CNS-1910193.
Portable DNA sequencers, such as Oxford Nanopore MinION, are fundamentally changing how DNA-based diagnostics is done. Instead of spending days in a lab, working with bulky bench-top machines, we can now take a pocket-size sequencer into the field, and do sequencing in-situ. In many cases, such capability is essential for, e.g., detecting and tracking spread of infectious diseases or metagenomic surveys in rapidly changing environments (see our HotMobile 2018 paper for more extensive discussion). When attached to mobile devices such as laptops or tablets, portable DNA sequencers provide in real-time signals describing detected DNA. However, the current software tools required to process and analyze these signals are not well suited to execute directly in the field, where energy, Internet connectivity, and compute power are extremely limited.
In this project, we aim to develop, and test in real-world use scenarios, new software system and algorithms to enable DNA processing directly on mobile devices, without the need to access advanced computational infrastructure.
Team
This project is a collaboration between the SCoRe Group and:
- Dr. Lauren Sassoubre (University of San Francisco and UB Department of Civil, Structural and Environmental Engineering),
- Dr. Ian Bradley (UB Department of Civil, Structural and Environmental Engineering).
Our Hardware
To run the project we are using two key pieces of technology. The first one is MinION sequencer from Oxford Nanopore Technology. The pictures below show one of the three devices we are using in our labs.
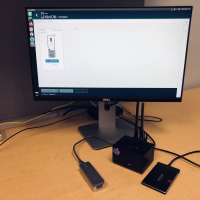
The second component is a Supercomputer On a Chip (SCoC). The specific platform we are using is based on NVIDIA's Jetson Nano. Below we provide pictures of the entire setup. We include also specification if you are interested in building a similar box for your project (please note that we are not endorsing any particular provider).
SCoC Harware Specification
- Jetson Nano – quad-core ARM Cortex-A57 processor, NVIDIA Maxwell with 128 GPU cores, 4 GB 64-bit LPDDR4 1600MHz RAM. Very powerful SCoC, running Ubuntu Linux:
18.04.3 LTS (GNU/Linux 4.9.140-tegra aarch64). - Geekworm NVIDIA Jetson Nano Metal Case – nice and small form factor package.
- NOCTUA NF-A4x20 PWM 5V fan – fan is critical to sustain Nano's operation under heavy load, it has to be 5V.
- Intel Dual Band Wireless-AC 8265 with 6dBi RP-SMA Dual Band 2.4GHz 5GHz Antennas – Nano board comes with standard GbEth port only. If you need WiFi connectivity, this solution requires almost no effort.
- Samsung 256GB U3 MicroSDXC EVO internal memory card – this is internal storage used by OS and software tools.
- WD 4TB Elements USB External Hard Drive – this is external storage to keep data (we recommend that you pick tested hard drive). Overall, a standard spinning hard drive is not optimal (it draws too much energy). When we do not need that much extra storage we use smaller SSDs.
- TOGUARD 7in 1024x600 TFT LCD display – we are not entirely happy with this option, you may consider a better solution.
- Pwr+ 5V Micro-USB charger – we use this particular type of charger a lot, no complaints.
Software Specification
Jeston Nano comes with its own variant of Ubuntu Linux for Tegra – the specific version is Ubuntu 18.04.4 LTS (GNU/Linux 4.9.140-tegra aarch64). Since this is fully functional Linux, we have access to all common tools, including compilers, like gcc and clang, and the entire NVIDIA CUDA development environment (currently we are using CUDA 10.0).
Of course, the key requirement is to be able to run Oxford Nanopore tools. ONT provides MinKNOW framework in its repository. Just running fairly standard chain of Debian/Ubuntu apt commands brings in all tools (notice that we are using xenial repository):
echo “deb http://mirror.oxfordnanoportal.com/apt xenial-stable-minit non-free” > /etc/apt/sources.list.d/nanoporetech.list
apt update && apt upgrade
apt install minknow-core-minit-offline ont-bream4-minit ont-configuration-customer-minit ont-minknow-frontend-minit ont-minknow-static-frontend
The only caveat is that currently, it is not possible to install guppy basecaller directly from packages (there are several reasons: first, dependencies that cannot be satisfied without downgrading Ubuntu, second, lack of support for NVIDIA SM53 in the default package). However, we were able to easily bypass these problems by compiling (with bit of editing) guppy from sources (we will share the process soon). Below is a glimpse at MinKNOW running on NVIDIA Nano.
We will be sharing our experiences and updating this web page as the project progresses!