GPU Computing
|
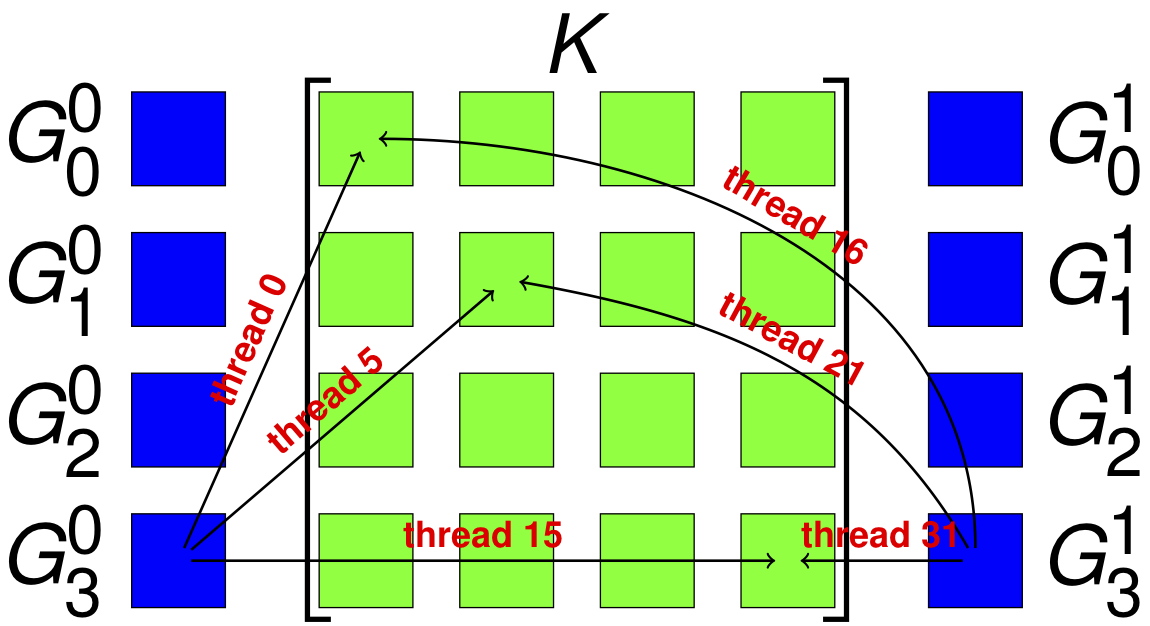
The combination of low power consumption, large memory bandwidth, high flop rate, and low cost make GPUs a promising platform for desktop scientific computing. The large latency is problematic for strong scaling, but there are many candidate computations for this type of throughput device, especially since interconect latency looks set to drop rapidly. Moreover, it is likely that a large fraction of future supercomputers will also employ accelerators (as 3 of the top ten machines in the world already do). In the last 4 decades of scientific computing, the most successful strategy for accommodating new hardware is the production of high quality libraries, such as MPI and PETSc. We propose to develop not only parallel algebraic solvers, but discretization and multiphysics libraries, suitable for hybrid machines with significant accelerator components. Our strategy is to use a combination of low-level library routines and code generation techniques to produce implementations for our higher level API. We use the Thrust, Cusp and CUSPARSE libraries from NVIDIA, and the ViennaCL libraries to provide linear algebra, shown here, and more basic operations such as sorting and keyed reductions. For operations that are more variable, such as those that depend on the weak form, we use code generation and dynamic compilation, as is available from OpenCL. The key point is to circumscribe the domain so that the generation process becomes tractable. In generating FEM integration routines, we produce not only different block sizes, but different organization of memory writes and computation. With Andy Terrel and Karl Rupp, we are exploring code transformations which will enable us to automatically optimize more general calculations. In particular, we are reorganizing traversal, and using code transformation tools, to explore quadrature-based methods for FEM residual and Jacobian evaluation. In our upcoming paper, we show that excellent vectorization is possible with CUDA and OpenCL, even for forms with variable coefficient. |
Papers
|
|